A tiny machine learning framework
Making MiniML, a small ML framework using Python and Jax
This summer I’ve had a couple of weeks on leave in a place of sun and sea; it stands only to reason of course that I’d spend a few of them creating my own new machine learning framework.
Did the world at large need one more? Probably not really. But I personally did, and if anyone else has any use for it, even better.
Why going “minimal”?
The new framework is called MiniML (pronounced minimal). My main reason for building it was that I’m generally interested in experimenting with small models and small problems, the kind that are easily tractable even on a laptop with a decent GPU. But I always felt caught between two options:
scikit-learn is fantastic to build modular pipelines (including tooling for processing data or adding features) and for training utilities, but its models are essentially “frozen in place”; it provides a lot of the classic models but they’re all precompiled binaries that the Python library wraps around, and that means beyond what their parameters allow they can’t really be modified;
PyTorch is very powerful and allows full customisation, but it’s often annoying in terms of how much boilerplate it requires to set up even for a basic training run, and its performance only shines when you start scaling up.
The thing I wanted was the power of PyTorch with the simplicity of scikit-learn. So I set up to do just that.
Making a difference
The core of a flexible machine learning framework has to be the engine providing the ability to automatically differentiate any function, to enable it to optimise the most diverse combinations of model and loss function. I considered trying making one myself as that’s probably the most educational bit of the work but have postponed that to a future exercise, and decided instead to rely on Jax. Jax is a framework that provides JIT compilation, automatic differentiation and GPU support for tensor operations, with a module that basically reproduces 1:1 the same interface as NumPy. I used Jax for all the automatic differentiation needs. For optimisation algorithms, instead, I used SciPy. I’m focusing on small models, so to begin with, minimisation algorithms like L-BFGS (provided by SciPy’s optimisation module) are perfectly fine and in fact probably far more performant than Adam or regular gradient descent; the main reason to use those is that when we have models with an extremely large number of weights, any algorithm that either computes or approximates the Hessian becomes completely unfeasible.
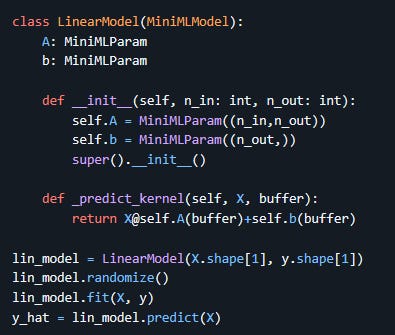
The core interface is drawn directly from PyTorch, with a few changes. Here is a basic example:
parameters are
MiniMLParamobjects, which specify the dimensions and optionally type and regularisation loss function for that parameter;models are
MiniMLModelobjects. They need to subclass from this but can in turn contain instances of other models;all child parameter and models must be direct members of the parent and declared in the constructor before the parent constructor is invoked. If you need to use lists of parameters or models, there are dedicated container classes for that;
each model also needs its
_predict_kernelmethod implemented, taking in a data array and a buffer and returning the computed result. This is where the core of the model is actually described, and this implementation should use Jax functions for all math. The buffer is where all the parameters are stored during training, and you only need it to pass it to the parameters (whose value is retrieved by simply calling the parameter object with the buffer as argument) or to any child models (invoked directly with their_predict_kernelmethod as well).
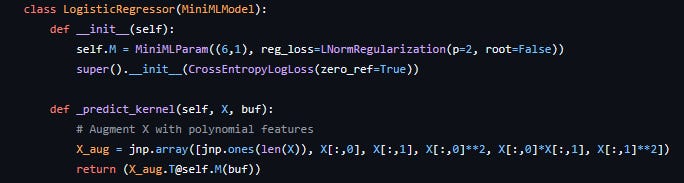
The model can then be initialised, randomised (this also creates the parameter buffer for the first time), and fit to data with a simple function call. You’ll notice here that no loss function was specified, which means the default was used (root mean square error). Different loss functions, including custom ones created by the user, can be passed to the parent constructor of a MiniMLModel. In addition the fit method takes in a regularisation lambda parameter to adjust the strength of regularisation, which is computed on each parameter using the function passed to them on creation. Here is for example an implementation of a custom logistic regression model for a specific problem with some extra polynomial features that makes use of these other functionality:
The model, of course, can also be saved and loaded from memory. See more in the documentation.
Other features
Other features provided:
a few basic implementations of simple common models, like a MLP and a Radial Basis Function layer, in the
miniml.nnmodule. Future plans include adding convolution layers and self-attention;a pre-fitting routine that allows you to fix or initialise some parameters by means other than loss minimisation. This is done by overloading the
_pre_fitmethod, and returning a set of any parameter names that should not be optimised any further;ability to get your parameters in dictionary form, and if necessary manually change them and set them back.
I plan to add more and I’ll make use of it for future posts here.